Introduction
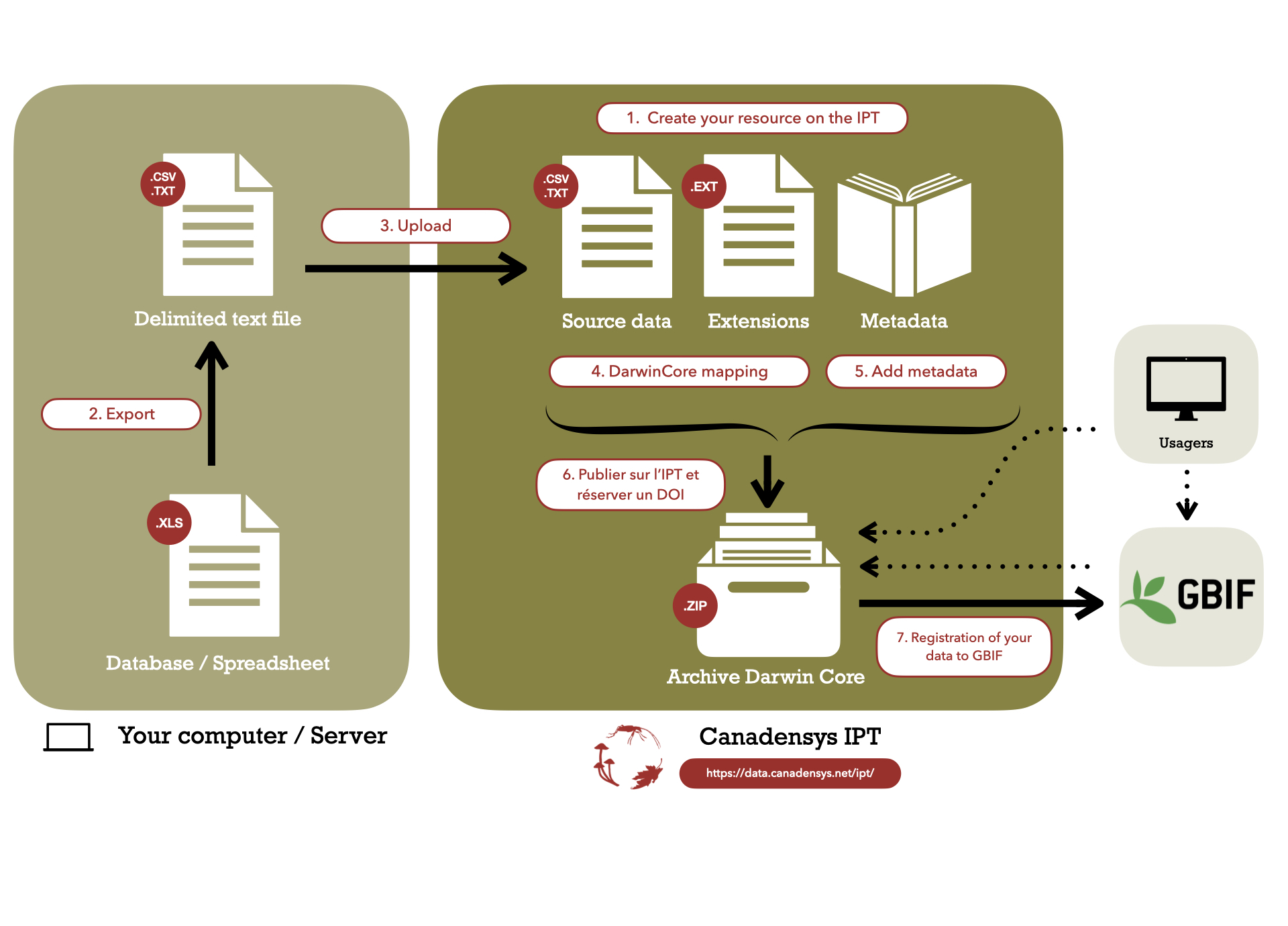
This guide explains how to publish your biodiversity data to GBIF using the Canadensys repository. The Canadensys repository is powered by the GBIF Integrated Publishing Toolkit (IPT) and customized by Canadensys to be fully integrated into our website. It allows you to upload, standardize, publish and register your data in 7 steps. Importantly, the data is published in your organisation’s name, for free.
Dataset format
Biodiversity data are published on Canadensys in the Darwin Core standard (DwC). It includes a glossary of defined terms (referred to dwc:termName herafter) and allows your data to be understood and used by anyone.
The Darwin Core Standard provides a robust and stable way to compile and share biodiversity data, regardless of the technology used to share it. It also allows the GBIF aggregator to integrate your data with other data worldwide.
It is not necessary to use these terms in your source data, but keeping them in mind should help you build the foundations of your database.
Classes of datasets
There are four classes of datasets supported by the IPT. Make sure to identify the one that is most appropriate for your biodiversity data.
For information about the different classes of datasets, see the following GBIF guides:
- Data quality requirements to publishing occurrence data
- Data quality requirements to publishing checklists
- Data quality requirements to publishing metadata
- Data quality requirements to publishing sampling-event data
Conditions
We care about data and we just want to make sure you do too. In order to publish your data using the Canadensys repository you should meet the following criteria:
- You are associated with a Canadian collection or organization.
- You are publishing one of the four types of datasets supported by the IPT.
- You hold the rights to publish the data.
- You are willing to maintain the dataset and improve its quality where possible.
- You are willing to provide sufficient metadata, so users can learn what the dataset is about.
- You are publishing the data under an open license, so others can really use them. We strongly recommend publishing under CC0.
Institution and collection registrations
Before we get started, it is essential to register your institution and collection on GRSciColl. The Global Registry of Scientific Collections (GRSciColl), is a comprehensive, community-curated repository of information about scientific collections. You will also need to register your institution or organization as a publisher on GBIF.
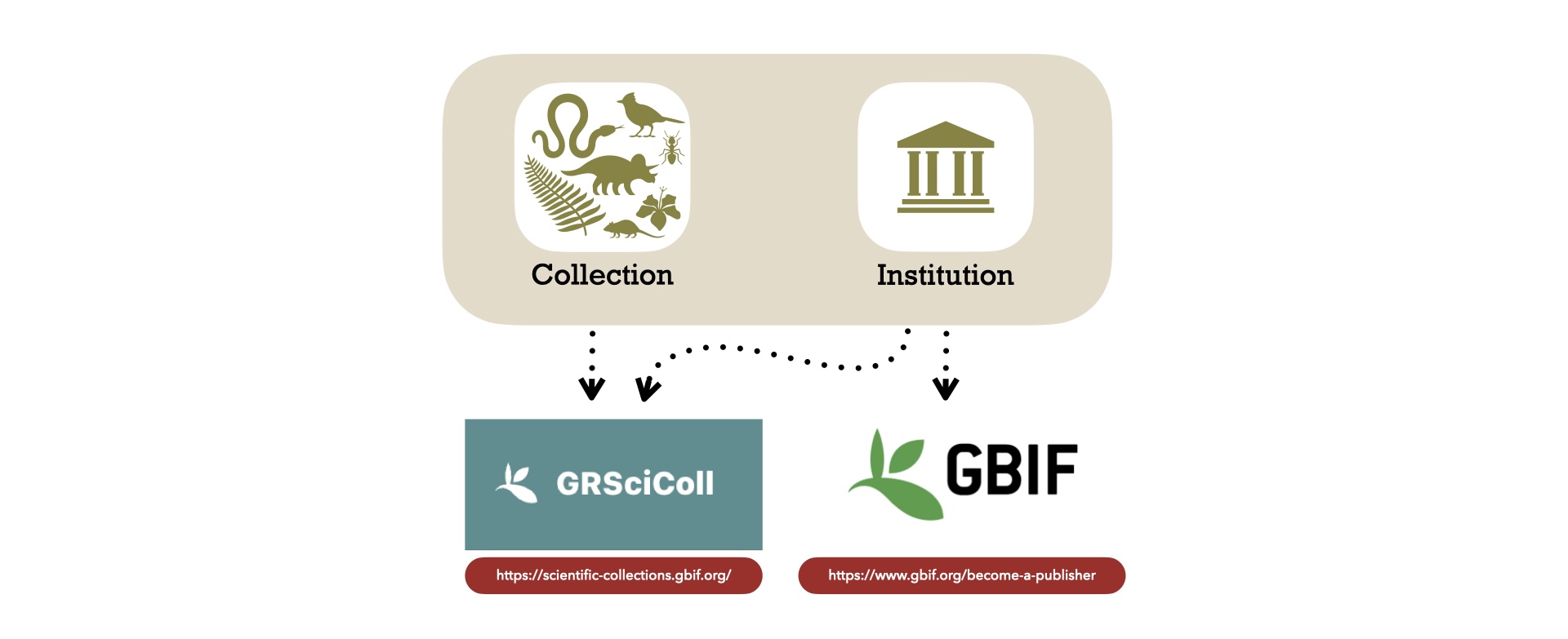
Add your instituion on GRSciColl
Add your collection on GRSciColl
1. Create your resource on the IPT
Let’s get started !
To be able to create and manage your own dataset (called a “resource”), you will need a user account. Simply contact us to create one for you and your team by providing the respective pertinent names and email addresses.
Once you have an account, login at the top of this page. Click on the tab Manage resources to get access to your dashboard. It will display all the datasets you are managing and will be empty at first. You can create a new resource at the bottom of the page. Follow the IPT manual for more detailed instructions.
NOTICE: Please use the following lowercase format for the shortname of your resource: yourCollectionCode-datasetType (e.g. acad-specimens or wildlife-sightings-observations). This name is used to uniquely identify and access your resource and cannot be modified subsequently! For testing purposes, please use yourCollectionCode-test (e.g. ubc-test).
Once you have created your resource, you will see an empty resource overview page.
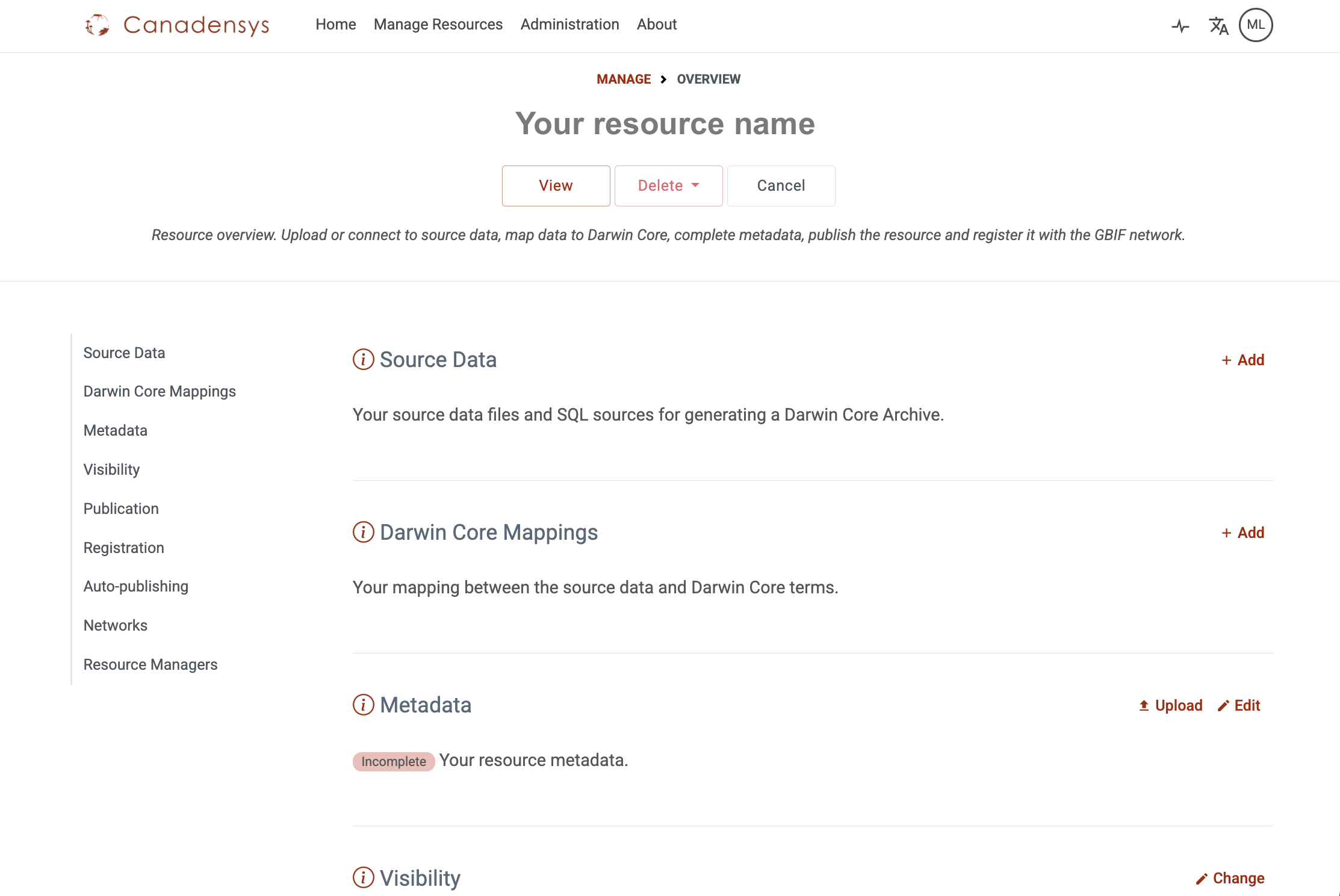
You can add the resource managers that can have access to your resource by adding them at the bottom of the resource page.

2. Export
Export your database as a delimited text file, This ensures that your data values are separated in each row with specific delimiter characters (e.g. .txt, .tab, .csv).
Examples: Delimited data formats have fields/columns separated by a character (tabulation character, coma, semicolon) and records/rows terminated by newlines.
specificEpithet,infraspecificEpithet,occurrenceRemarks
Leucoagaricus,naucinus,"Some text, with a coma"specificEpithet;infraspecificEpithet;occurrenceRemarks
Leucoagaricus;naucinus;"Some text; with a semicolon"specificEpithet infraspecificEpithet occurrenceRemarks
Leucoagaricus naucinus "Some text, with a coma"Encoding assignation can also sometimes make things tricky because of misinterpretations of accented characters (e.g. é, à, ü, î) or the degree character (°). This is especially the case in authorship information (dwc:scientificNameAuthorship), the person/group/organization who assigned the Taxon name (dwc:identfiedBy), and the geographic coordinates (dwc:decimalLatitude and dwc:decimalLongitude).
Select the UTF-8 character encoding for your export. If the option is available, include a header line in your export (a first line with the field names), as it will be helpful later.
WARNING: Please avoid using the character incoding ASCII, Macintosh, and Windows ANSI.
At this step, you can contact us if you want to get a quick check of your data before uploading it to the IPT.
3. Upload
To import your data (your source data, and optionally one or several extensions), go to your resource overview page > Source Data and click Add. You can import new data, or you can also import your source data as an already archived resource (Darwin Core Archive, zipped IPT resource configuration folder, or metadata file). You can find more information about the source files you can import here.

NOTICE: You might want to compress/zip your source file first to improve the upload speed of large files. The IPT will unzip them automatically once received. Follow the IPT manual for more detailed instructions, including the option to use multiple source files or to upload via a direct database connection.
It is possible to map an extension at this step, as long as it is different from the core type mapped.
Once your source file has been uploaded correctly, a source file detail page will be shown (see an example screenshot in the IPT manual), displaying how the IPT has interpreted your file (number of columns, rows, header rows, character encoding, delimiters, etc.). Click on the Edit button of your source file, and click Options > Preview button to verify everything is correct, then click Save.
4. Darwin Core Mapping
Darwin Core mapping is the process of linking the fields in your source file with the appropriate Darwin Core terms.
If your source data already uses Darwin Core terms in the header of your data, the mapping process is completely automated. A simple verification of the mapping is then necessary to validate this step. Once verified click on the Save button.
If your data do not use Darwin Core terms, this step can be challenging for two reasons:
- The list of Darwin Core terms can be overwhelming, so it might be difficult to select the ones that are appropriate for your dataset.
- The IPT currently only allows one-to-one mapping of fields, so the ease of mapping will depend on your database structure and on the feasibility of exporting as close to Darwin Core as possible.
This is why we are here to help! Contact us to guide you through the steps, review your mapping, suggest terms and help you repeat steps 2-4 until the mapping is just right.
You can find more information regarding Darwin Core mapping in the IPT manual (including core types, extensions, auto-mapping, default values, value translation, etc.) and in the introduction to Darwin Core on our website.
5. Add metadata
If data are LEGO bricks, then metadata are the shiny box and instructions. They enable users to discover your dataset and assess its relevance for their particular needs, so it pays off investing some time providing them.
Go to your resource overview page > Metadata and click Edit to open the metadata editor. Link your resource with your institution in the metadata once it is registered to GBIF (see this section in the introduction of this page).

Any information you provide here will be visible on the resource homepage and bundled together with your data when you publish. Metadata are expressed in the GBIF EML Profile standard
NOTICE: The GBIF EML Profile can also be downloaded as a Rich Text Format (RTF) file using the Upload button. The latter can serve as a draft manuscript describing the dataset (a “Data Paper“), which can be submitted for peer-review to a Pensoft open-access journal such as the Biodiversity Data Journal, Phytokeys, Zookeys, Biorisk, Neobiota or Nature Conservation.
Follow the IPT manual for detailed instructions about the metadata editor and use one of the currently published datasets as an example:
6. Publish
By default, the visibility of your resource is set on private. Once you are ready to make it available to everyone, go to the Visibility section, and click on Change to set it to Public. See the IPT manual for more information.

To publish your dataset, click on Publish. You can also publish while the Visibility is on private. This can allow you the save the versionning of your dataset among the managers of the resource before it is ready to share publicly. When you decide that the published version is satisfactory, you can change the Visibility to Public.
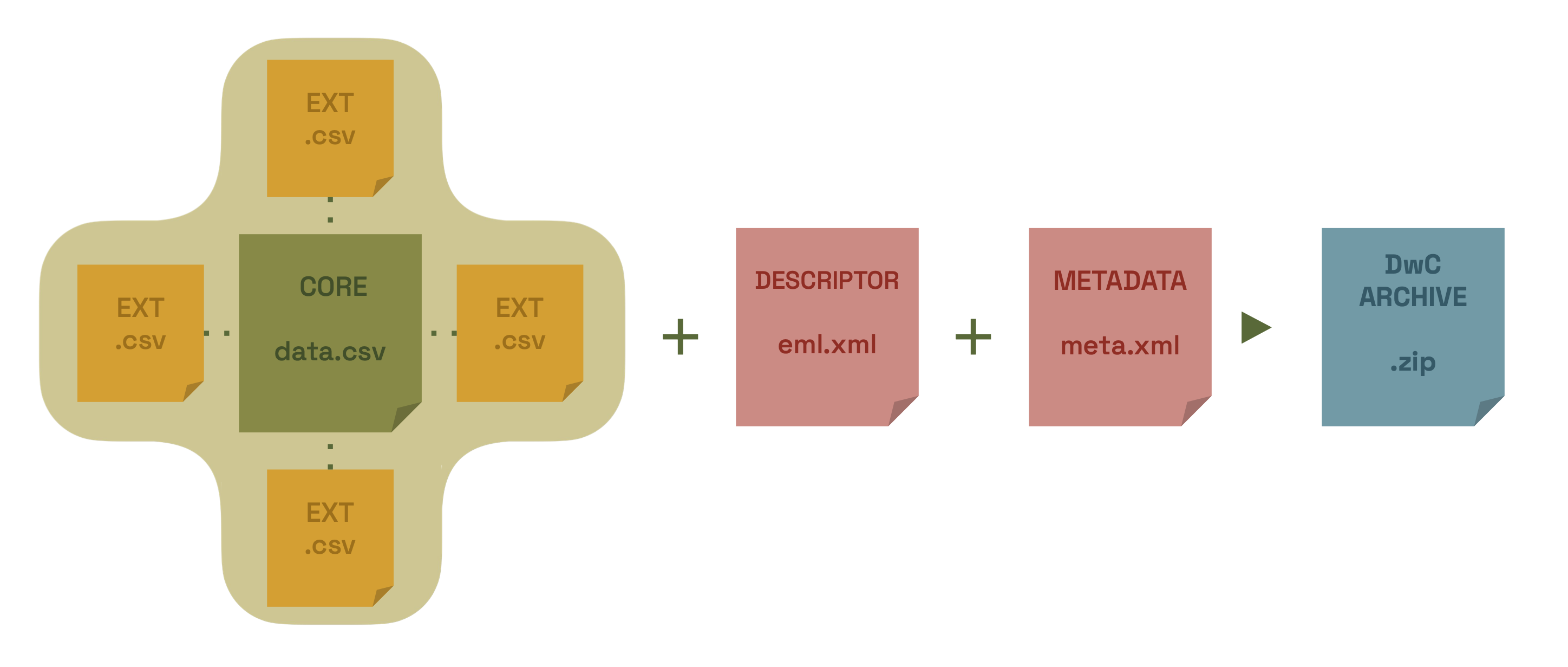
During the publication process, the Canadensys IPT will generate your data as Darwin Core, combine it with the metadata and package it as a standardized zip-file called a Darwin Core Archive.
If you wish to assign a DOI to your dataset, you need to click on the Reserve button  , then publish again using the Publish button.
The DOI is registered directly with DataCite and a link will be added in the metadata of your resource. If your dataset has not been published yet, you have to publish it first, reserve a DOI, and publish again. The version will then switch from version 1.x to 2.0 as it is a major change in your dataset metada.
, then publish again using the Publish button.
The DOI is registered directly with DataCite and a link will be added in the metadata of your resource. If your dataset has not been published yet, you have to publish it first, reserve a DOI, and publish again. The version will then switch from version 1.x to 2.0 as it is a major change in your dataset metada.
For more information about the DOI assignation steps, see the IPT DOI Workflows. See the IPT manual for more details about the general publication process.
NOTICE: Do not publish publicly any test dataset if you use one.
Congratulations! You just published your dataset to the world! It is now listed on the repository homepage and you can share its link using this link format and your dataset shorname:
https://data.canadensys.net/ipt/resource?r=dataset-shortnameYou can now notify any regional or thematic network you are involved in, such as VertNet, the Consortium of Northeastern Herbaria or the Entomological Society of Canada.
Keep in mind that your published dataset is a static snapshot of your data and will not change until you upload an updated source file and click publish again. This procedure has the advantage that your dataset is always available, does not require a live internet connection to your database and can be easily shared (e.g. you can email the Darwin Core Archive to a colleague). It also allows you to control the publication process more precisely (version 1, version 2, etc.) and users are informed of how recent the data are and the differences between versions (e.g. addition of data, correction of errors, modification of metadata, and so on).
7. Register with GBIF
Even though your dataset is now available to everyone on the Canadensys IPT, you can enhance its accessibility by users by registering your dataset with the Global Biodiversity Information Facility (GBIF). It allows your data to become available to an international audience via the GBIF portal. It also ensures full attribution is given to your institution. By registering your data, you agree with the GBIF Data Publisher Agreement.
On the resource overview page, in the registration section, click on Register. You can then confirm that you are in accord with the GBIF data sharing agreement in the dialog box. Your dataset is now available on GBIF! It will allow GBIF to index your resource on their portal, where it can be easily accessed by everyone. See the IPT manual) for more details about the registration step.

NOTICE: If your dataset has been already published (either entirely or partially) on GBIF via an another IPT, measures needs to be taken to link the old and new dataset to avoir duplicates. Contact us so we can arrange this additional step in the publication process!
Citation
As all documents on this website, this guide is published under CC-BY.
To cite this guide, use the citation:
Desmet, P.; Leménager, M.; Sinou, C. 2025. 7-step guide to data publication. Canadensys. https://canadensys.net/publish/7-step-guide/